Abstract
Background: Diffuse large B-cell Lymphoma (DLBCL) is stratified histologically as GCB and ABC phenotypes based on Hans algorithm. Besides this categorization, there is a paucity of molecular markers useful for predicting response to CHOP/R-CHOP chemotherapy and outcome of DLBCL patients. We performed miRNA profiling to identify potential miRNAs for their ability to predict response to chemotherapy and outcome in DLBCL patients.
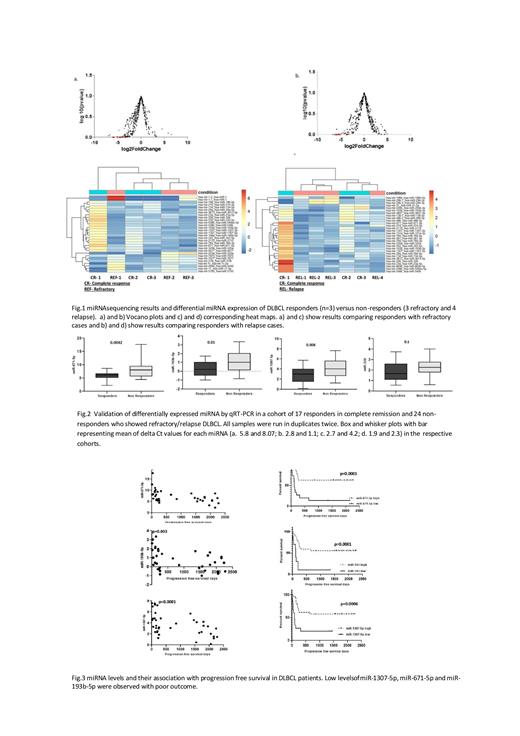
Methods: A total of 41 cases of histologically and immunohistochemically confirmed DLBCL cases who received R-CHOP/CHOP were included in this study after IRB approval. The follow up ranged from 7 to 2273 days with a mean of, 874 days. Cases in sustained complete remission for ≥ 1095 days were identified as "responders" whereas cases with partial response/stable disease or primary progressive disease were identified as"non-responders". In the first part of the study, miRNA transcriptome profiling was performed from lymph node tissue of 10 cases of DLBCL which included 3 responders and 7 non-responders (3 with refractory disease and 4 with relapse) for identification and selection of differentially expressed miRNAs. Total RNA was isolated from the biopsies, libraries constructed with ION TOTAL RNA-SEQ v2 and sequenced on S5 ION Torrent. The filtered reads were aligned to the reference genome hg19 using Bowtie. Differentially expressed miRNAs were identified by DESeq2. In the second part of the study, validation of the selected miRNAs was performed in a retrospective validation cohort of 41 cases including 17 responders and 24 non-responders by semi-quantitative qRT-PCR. ROC curves were obtained and best cut-off for the delta Ct values was selected for each of the miRNAs to distinguish the two cohorts of patients. In the third part of the study, an additional 38 cases of DLBCL were included prospectively as testing cohort, in whom mi-RNA were isolated from paired pre-treatment biopsy and plasma samples were evaluated for the expression of the selected miRNAs for prediction of response to treatment and outcome.
Results: miRNA transcriptome profiling revealed a total of 30 differentially expressed miRNAs among DLBCL responders vs non-responders and the data was submitted to GEO database (GSE179760). The top 4 significantly downregulated miRNAs in non-responders selected for validation included miR-193b-5p, miR-671-5p, miR-1307-5p and miR-326 as shown in the volcano plot and heatmap in Fig.1. In-silico prediction for the interacting genes of the selected miRNAs revealed genes involved in lymphoma progression and chemoresistance . Validation by qRT-PCR revealed significant down-regulation of miR-1307-5p, miR-671-5p and miR-193b-5p in the non-responders compared to responders (Fig.2). There was no significant association of miRNA expression with disease stage, nodal or extranodal origin and the presence of B symptoms. However, the downregulation of miR-671-5p was found to be associated with the presence of B-symptoms. Downregulation of miR-193b-5p, miR-671-5p, miR-1307-5p expression showed a significant association with progression free survival as shown in Fig.3 (Spearman's correlation plot and Kaplan-Meier survival curves).
In the prospective cohort of 38 DLBCL patients, all 3 miRNAs were expressed in cell free plasma and tissue but only miRNA-193b-5p showed correlation between them. Using the cut-off Ct values obtained from ROC curves of tissue miRNA levels in the validation cohort , 16/38 patients were predicted to be non-responders. The preliminary data showed that 8 of these 16 cases were correctly predicted (6 died of disease, 2 showed partial response). The remaining cases are still under follow-up. The treatment response could not be predicted on the corresponding plasma levels.
Conclusion: Downregulation of miRNA-671-5p, miR-193b-5p and miR-1307-5p in pre-treatment biopsy samples is associated with poor outcome and shorter progression free survival and poor treatment response leading to refractory/relapse DLBCL, which needs confirmation in larger studies.
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal